Photoactive Yellow Protein
By Jocelyne Vreede
How can a light signal, very small in size en very short in time, lead to a long-lasting, organism-wide response?
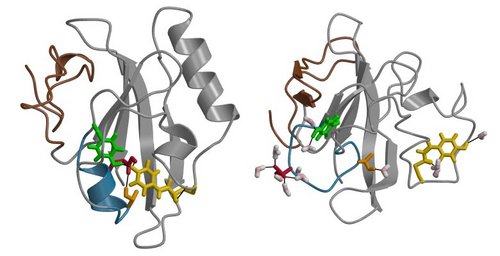
To investigate (part of) this question, we study the Photoactive Yellow Protein (PYP) from the bacterium Halorhodospira halophila, where it is involved in a negative phototactile response to blue light. Comprising 125 amino acids and a covalently bound chromophore, the protein folds into a alpha-beta core capped by an N-terminal domain, containing two helices. Upon absorbing a blue-light photon as a trigger, PYP undergoes a photo cycle, starting from its receptor state. Visiting several intermediate states, the chromophore twists along its double bond to a cis configuration within picoseconds. Within microseconds of the isomerization, a proton from Glu46 (protonated in the receptor state) transfers to the chromophore, leaving a negative charge on Glu46 (see also the research of Elske). Driven by the new negative charge in the chromophore binding pocket, the protein subsequently unfolds to expose the chromophore and Glu46 to bulk water forming the signaling state. The completion of the photo cycle, i.e. the refolding of the protein to the receptor state pG, requires several hundreds of milliseconds.
We study the conformational transitions in PYP in atomic detail, using molecular simulation methods, including parallel tempering, metadynamics and transition path sampling (see also the research of Jarek). So far, we have been able to predict conformational characteristics of the signaling state of PYP and a dynamical bottleneck in the recovery of the receptor state.
See also: the illustrative movie on our gallery webpage.
Publications
- Predicting the signaling state of Photoactive Yellow Protein. Biophys J. 88 (2005), 3525 DOI: 10.1529/biophysj.104.055103
